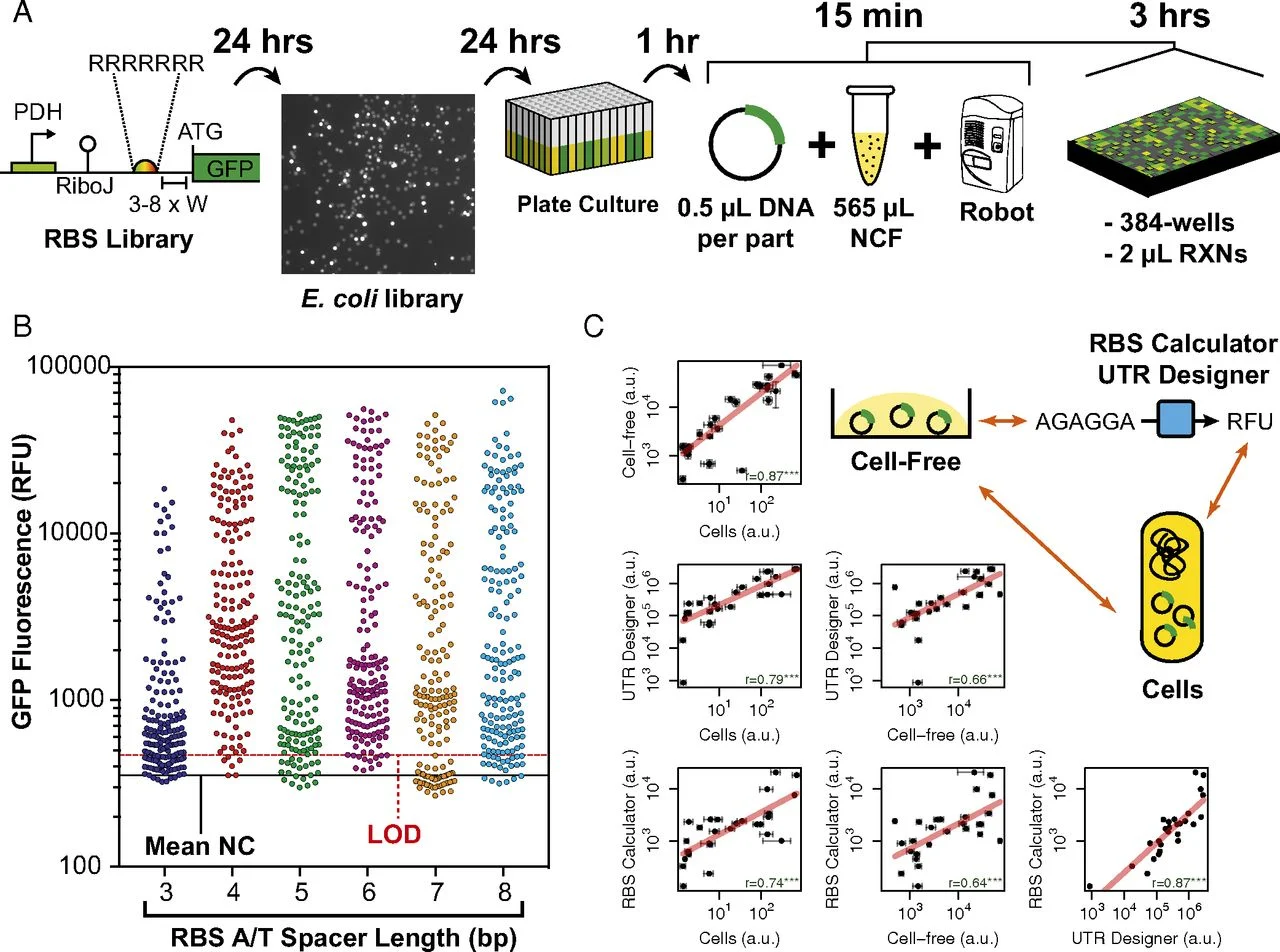
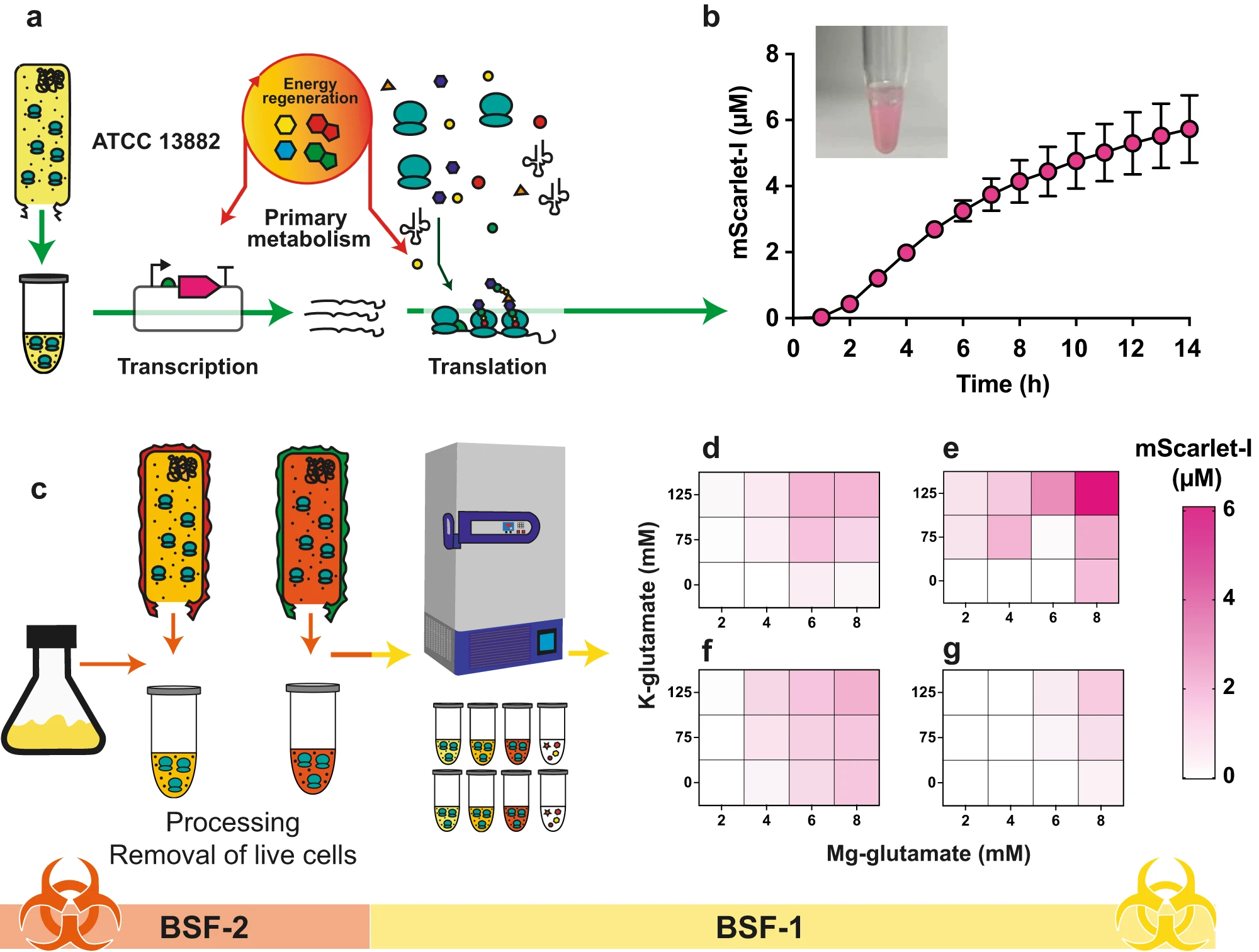
Microbes harbour extraordinary potential to produce complex chemicals encoded within biosynthetic gene clusters (BGCs), yet most remain silent or inactive under laboratory conditions. Our research addresses fundamental questions about pathway regulation and biosynthesis while also developing strategies to unlock silent BGCs. While basic science has a prominent role in our approach, we consider how research can be applied to global challenges including antimicrobial resistance, sustainable crop protection, and replacing petrochemical synthesis with biological alternatives. We specialise in engineering Streptomyces venezuelae, a fast-growing synthetic biology model, to discover and produce novel bioactive compounds.
Our basic research focuses on understanding mechanistic questions such as pathway regulation, single enzyme characterisation all the way through to complex multistep biosynthetic pathways. This fundamental knowledge also enables applied outcomes including enzyme discovery, pathway engineering, and production of high-value chemicals and novel biotherapeutics. We employ synthetic biology tools including CRISPR-Cas9, heterologous expression hosts, and pathway refactoring to both elucidate natural biosynthetic mechanisms and unlock nature's hidden chemical diversity for therapeutic and industrial translation.